Difference between revisions of "SMT profiling with pmcstat and perf"
| (50 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | This article discusses profiling symmetric multithreading (SMT) on the POWER9 architecture. It uses both | + | This article discusses profiling symmetric multithreading (SMT) on the POWER9 architecture. It uses both Big-endian FreeBSD with pmcstat and Debian with perf. |
| + | |||
| + | The knowledge presented here was derived from a variety of sources which can be found in the [[#Additional Resources]] section. | ||
| + | |||
=== Symmetric multithreading (SMT) === | === Symmetric multithreading (SMT) === | ||
| Line 5: | Line 8: | ||
==== SMT principles ==== | ==== SMT principles ==== | ||
| − | ===== SMT is not "multi-core" ===== | + | ===== SMT is "multi-thread" not "multi-core" ===== |
| + | This is an important distinction. SMT is a technology that increases throughput of instructions through parallelization where there are under-used CPU components. While SMT4 can support four threads per core and SMT8 can support eight threads per core, this is not an additional three and seven cores, respectively. There are trade-offs and benefits. Per-thread performance declines with increasing utilization of SMT levels, but overall performance and power consumption efficiency increase. Note that IBM did not market SMT as "multi-core," while several media sites conflated SMT with increased core count. | ||
==== Comparison to RISC-V HARTs ==== | ==== Comparison to RISC-V HARTs ==== | ||
| + | |||
| + | === Operating Environment === | ||
| + | ==== Hardware ==== | ||
| + | The bare metal hardware is an eight core SMT4-capable system from Raptor. | ||
| + | |||
| + | <pre> | ||
| + | # lscpu | ||
| + | Architecture: ppc64le | ||
| + | Byte Order: Little Endian | ||
| + | CPU(s): 32 | ||
| + | On-line CPU(s) list: 0-31 | ||
| + | Model name: POWER9 (raw), altivec supported | ||
| + | Model: 2.3 (pvr 004e 1203) | ||
| + | Thread(s) per core: 4 | ||
| + | Core(s) per socket: 8 | ||
| + | Socket(s): 1 | ||
| + | Frequency boost: enabled | ||
| + | CPU(s) scaling MHz: 67% | ||
| + | CPU max MHz: 3800.0000 | ||
| + | CPU min MHz: 2166.0000 | ||
| + | Caches (sum of all): | ||
| + | L1d: 256 KiB (8 instances) | ||
| + | L1i: 256 KiB (8 instances) | ||
| + | NUMA: | ||
| + | NUMA node(s): 1 | ||
| + | NUMA node0 CPU(s): 0-31 | ||
| + | Vulnerabilities: | ||
| + | Gather data sampling: Not affected | ||
| + | Indirect target selection: Not affected | ||
| + | Itlb multihit: Not affected | ||
| + | L1tf: Mitigation; RFI Flush, L1D private per thread | ||
| + | Mds: Not affected | ||
| + | Meltdown: Mitigation; RFI Flush, L1D private per thread | ||
| + | Mmio stale data: Not affected | ||
| + | Reg file data sampling: Not affected | ||
| + | Retbleed: Not affected | ||
| + | Spec rstack overflow: Not affected | ||
| + | Spec store bypass: Mitigation; Kernel entry/exit barrier (eieio) | ||
| + | Spectre v1: Mitigation; __user pointer sanitization, ori31 speculation barrier enabled | ||
| + | Spectre v2: Mitigation; Software count cache flush (hardware accelerated), Software link stack flush | ||
| + | Srbds: Not affected | ||
| + | Tsx async abort: Not affected | ||
| + | |||
| + | # lsmem | ||
| + | RANGE SIZE STATE REMOVABLE BLOCK | ||
| + | 0x0000000000000000-0x00000007ffffffff 32G online yes 0-31 | ||
| + | |||
| + | Memory block size: 1G | ||
| + | Total online memory: 32G | ||
| + | Total offline memory: 0B | ||
| + | |||
| + | # facter | grep virtual | ||
| + | is_virtual => false | ||
| + | virtual => physical | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | ==== Software ==== | ||
| + | Three environments will be used for the operating environment. The bare metal runs Debian: | ||
| + | |||
| + | <pre> | ||
| + | # uname -a | ||
| + | Linux 93-224-155-23 6.1.0-37-powerpc64le #1 SMP Debian 6.1.140-1 (2025-05-22) ppc64le GNU/Linux | ||
| + | </pre> | ||
| + | |||
| + | Two QEMU VMs provide little-endian Debian and big-endian FreeBSD environments: | ||
| + | |||
| + | <pre> | ||
| + | # lscpu | ||
| + | Architecture: ppc64le | ||
| + | Byte Order: Little Endian | ||
| + | CPU(s): 16 | ||
| + | On-line CPU(s) list: 0,4,8,12 | ||
| + | Off-line CPU(s) list: 1-3,5-7,9-11,13-15 | ||
| + | Model name: POWER9 (architected), altivec supported | ||
| + | Model: 2.3 (pvr 004e 1203) | ||
| + | Thread(s) per core: 1 | ||
| + | Core(s) per socket: 4 | ||
| + | Socket(s): 1 | ||
| + | Virtualization features: | ||
| + | Hypervisor vendor: KVM | ||
| + | Virtualization type: para | ||
| + | Caches (sum of all): | ||
| + | L1d: 128 KiB (4 instances) | ||
| + | L1i: 128 KiB (4 instances) | ||
| + | NUMA: | ||
| + | NUMA node(s): 1 | ||
| + | NUMA node0 CPU(s): 0,4,8,12 | ||
| + | Vulnerabilities: | ||
| + | Gather data sampling: Not affected | ||
| + | Indirect target selection: Not affected | ||
| + | Itlb multihit: Not affected | ||
| + | L1tf: Mitigation; RFI Flush, L1D private per thread | ||
| + | Mds: Not affected | ||
| + | Meltdown: Mitigation; RFI Flush, L1D private per thread | ||
| + | Mmio stale data: Not affected | ||
| + | Reg file data sampling: Not affected | ||
| + | Retbleed: Not affected | ||
| + | Spec rstack overflow: Not affected | ||
| + | Spec store bypass: Mitigation; Kernel entry/exit barrier (eieio) | ||
| + | Spectre v1: Mitigation; __user pointer sanitization, ori31 speculation barrier enabled | ||
| + | Spectre v2: Mitigation; Software count cache flush (hardware accelerated), Software link stack flush | ||
| + | Srbds: Not affected | ||
| + | Tsx async abort: Not affected | ||
| + | |||
| + | # lsmem | ||
| + | RANGE SIZE STATE REMOVABLE BLOCK | ||
| + | 0x0000000000000000-0x00000001ffffffff 8G online yes 0-511 | ||
| + | |||
| + | Memory block size: 16M | ||
| + | Total online memory: 8G | ||
| + | Total offline memory: 0B | ||
| + | </pre> | ||
| + | |||
=== Benchmark code === | === Benchmark code === | ||
| + | ==== Description of comparison code (cmp_mp) ==== | ||
| + | The code being profiled and used for benchmarking is the genomic comparison code that the [https://mpjanson.org M. P. Janson Institute for Analytical Medicine] uses to look for molecular mimicry between pathogens and human tissue and hormones. This code uses a variety of techniques to compare nucleotide and amino acid sequences. There are both byte-by-byte and vector versions. The raw source code can be found at [[TBD]] | ||
| + | |||
| + | Fundamentally the algorithm just compares characters in two sequences for matching. The goal is to match consecutive characters between two sequences with constantly changing characters, and record the string and location of the matches. Unlike strstr or similar functions, the string to be matched is not known in advance. The first character in one sequence is compared against the first character in the other sequence. If it matches, the second characters are compared. If they match, the third characters are compared, and so on, until the characters do not match. If the length of the matched string is below the minimum desired length, the match is discarded and the comparisons start over at the next character. This can "slide" the comparisons relative to each other, so if the first four characters match but not the fifth, the comparison starts over at the first character of one sequence and the fifth character of the other. If there is a match of sufficient length, that two-dimensional region is marked as an "exclusion region" and not searched again. | ||
| + | |||
| + | The code can run N number of user-specified pthreads in parallel (-j N), by slicing one of the two sequences into smaller pieces and stitching the results together. N must be an even number (one forward, one reverse). Some matches are lost at the boundaries between slices when one or both parts are less than the minimum length. The execution time is extremely sensitive to comparisons made; adding checks to determine if a match falls on a slice boundary adds significant overhead. For large sequences, the odds of a matching string falling on a boundary are very low; for small sequences, running fewer threads is effective without costing significant time delays. For absolute accuracy, run only two threads (one forward and one in reverse). | ||
| + | |||
| + | ==== Description of data ==== | ||
| + | Nucleotides are denoted as A, T, C, and G. There are three nucleotides in an amino acid, and 23 amino acids in humans, which leads to "isocodon" amino acids. These have the same function but different nucleotide combinations. There are amino acids that denote the initiation and termination of a gene. The amino acid is determined by its offset from the initiation amino acid (ATG, methionine), which starts an "Open Reading Frame (ORF)." ORFs can overlap. DNA has a forward ("sense") and backward ("anti-sense") direction, so there are three possible amino acids reading forward, and three when reading backwards. Nucleotide sequences are usually published in the forward direction, and the other strand's nucleotide matching the forward direction is inferred (A matches T, and C matches G, and vice versa). Genome sequences can be obtained from the [https://www.ncbi.nlm.nih.gov/gene/ NIH National Library of Medicine] gene database, as well as in other places. The particular sequence is identified by its "accession number," and sequencing of the same organism by different labs does lead to different accession numbers. Some sequences are "canonical," meaning the generally accepted sequence that is often an assembly of accessions, and others are "contributions," meaning independently obtains sequences submitted for recording. | ||
| + | |||
| + | A typical "FASTA" file contains data as | ||
| + | |||
| + | <pre> | ||
| + | TATATTAGAGTAGGAGCTAGAAAATCAGCACCTTTAATTGAATTGTGCGTGGATGAGGCTGGTTCTAAATCACCCATTCAGTACATCGATATCGGTAATTATACAGTT... | ||
| + | </pre> | ||
| + | |||
| + | The most common comparison method is BLAST, which uses a database of pre-identified nucleotide sequences. This is useful for rapidly finding large matches, but does not offer granularity for exploring other aspects of the genome. The above method allows matching of nucleotide and amino acid sequences on both position one and position two nucleotide. This often reveals "repeat motifs" where a sequence of nucleotides is repeated multiple times within a pathogen. The current thinking is that this is an "immuno-evasive strategy," allowing the pathogen to escape detection by the host's immune system. Additionally, by recording the location of the matches, the distance between matches can be calculated. This is important in deciding if a host antibody would identify healthy tissue and hormones as pathogenic, by having similar shapes. The above algorithm also allows for detecting "SIDs," which are "Substitutions, Insertions, and Deletions," which occur regularly in rapidly evolving pathogens such as viruses. | ||
| + | |||
| + | For example, relative to the example above | ||
| + | |||
| + | <pre> | ||
| + | TTATATTAGAGTAGGAGCTAGAAAATCAGCACCTTTAATTGAATTGTGCGTGGATGAGGCTGGTTCTAAATCACCCATTCAGTACATCGATATCGGTAATTATACAGTT... | ||
| + | </pre> | ||
| + | |||
| + | is identical except for the inserted nucleotide T at the very first character. If the ORF is prior to the sequence above, this changes the amino acid structure by altering the anchor point for examining three nucleotides at a time (a "codon"). A straight matching of amino acids would miss this. The comparison code (cmp_mp) has the ability to shift each nucleotide sequence by none, one and two positions to catch insertions that alter the identifiable amino acids, and can match against the first character ("T" in "TAA") or the second nucleotide ("A" in "TAA"), which is more highly conserved across evolutionary pathways. | ||
| + | |||
| + | ==== Sample output ==== | ||
| + | |||
| + | The output is a csv file: | ||
| + | |||
| + | <pre> | ||
| + | CVD_OM003364.1,CVD_OM002793.1,225,2848,29762,29762,1,1, | ||
| + | 1,1,163,163,164, | ||
| + | ... | ||
| + | </pre> | ||
| + | |||
| + | The first line indicates the sequences compared, the number of matches, the longest match, the length of each sequence, and the presentation form, in this case a 1x1 image. The second line indicates a match was found from (1, 163) in one sequence and (1, 163) in the other, for a length of 164 characters, inclusive. | ||
| + | |||
| + | The csv file is run through some custom image generation code using libjpeg. An example of the processed data is below | ||
| + | |||
| + | <center>[[file:Mp_Bmiyamotoi_CP006647.2_vs_Bburgdorferi_GCF_000171735.2_20_1_100_nuc.gif]]</center> | ||
| + | |||
| + | The image shows matching sequences between two bacteria, [https://www.ncbi.nlm.nih.gov/nuccore?term=GCF_000171735.2 B. burgdorferi], which causes Lyme disease, and [https://www.ncbi.nlm.nih.gov/nuccore/CP006647.2/ B. miyamotoi], a bacteria in the same Borrelia family. Matches are occurring in both the forward and reverse directions. In this particular case, the B. miyamotoi sequence was assembled from fragments in both the "sense" and "antisense" direction. This is an artifact of how PCR is done, but has to be considered when doing analysis. Running only forward matching can lead to missed matches (a "false negative"). This comparison is particularly important because some people will exhibit symptoms of Lyme disease but only test weakly positive or not at all for Lyme disease. This is known in science but not practiced in medicine, leading to erroneous conclusions about the cause of the patient's symptoms (see [https://doi.org/10.1016/j.cmi.2019.07.026 Borrelia miyamotoi infection leads to cross-reactive antibodies to the C6 peptide in mice and men]). | ||
| + | |||
| + | The B. burgdefori sequence is 1,455,375 nucleotides long, and the B. miyamotoi sequence is 907,293 nucleotides long. Each nucleotide is compared at least once, and when there are partial matches too short to place the nucleotide into an exclusion region, it may be compared more than once. In this case, more than 2.6 trillion comparisons are made (one comparison is forward against forward, and one is forward against reverse; reverse against reverse is the same as forward against forward because of the inverse nucleotide arrangement of DNA). When searching human genes, the comparisons can be on the order of ''quadrillions'' (10^15) and take hours even on fast CPUs even with multi-core threading. Optimizing the comparison code saves time and power consumption, and is the focus of the profiling done below. | ||
| + | |||
| + | ==== Baseline comparison ==== | ||
| + | For these benchmarks, two different sequences of Epstein-Barr (EBV) and SARS-CoV-2 (Covid-19) virii will be used. EBV is the virus that causes mononucleosis, but is also closely associated with multiple sclerosis and has identifiable molecular mimicry sequences. SARS-CoV-2, the virus that causes Covid-19, evolved from a coronavirus, as can be shown by a comparison of amino acids (nucleotide shifts of none for [https://www.ncbi.nlm.nih.gov/nuccore/KU131570 CRNA] and one for [https://www.ncbi.nlm.nih.gov/nuccore/OM002793.1 Covid-19], anchor on second position, match initiation, termination and arginine amino acids): | ||
| + | |||
| + | <center>[[file:CVD_OM002793.1_vs_CRNA_KU131570.1_2_12_0_100_R2.gif]]</center> | ||
| + | |||
| + | The COVID-19 sequences, [https://www.ncbi.nlm.nih.gov/nuccore/OM002793.1 OM002793.1] and [https://www.ncbi.nlm.nih.gov/nuccore/OM003364.1 OM003364.1] are suitable for short duration benchmarks. The EBV sequences, [https://www.ncbi.nlm.nih.gov/nuccore/KC207814.1 KC207814.1] and [https://www.ncbi.nlm.nih.gov/nuccore/NC_007605.1 NC_007605.1] (canonical), are for longer benchmarks in which slight performance gains are measured. Each pair member in the two pairs of sequences is nearly identical to the other, so when analysis of branch prediction for misses is needed, one from each pair can be compared against the other. | ||
| + | |||
| + | A typical comparison command looks like | ||
| + | |||
| + | <pre> | ||
| + | ./cmp_mp -l 12 -j 8 -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | </pre> | ||
| + | |||
| + | It is specifying a minimum length of 12 consecutive nucleotide matches and four forward slices and four reverse slices, with the two Covid-19 sequences. For benchmark clarity, the -s (silence) flag is set. The output from BE FreeBSD for only two threads: | ||
| + | |||
| + | <pre> | ||
| + | $ time ./cmp_mp -l 12 -j 1 -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 8 | ||
| + | -l optarg = 12 | ||
| + | Number of jobs must be even; adding +1 to n_jobs | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764 | ||
| + | number of sequences: 222 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv | ||
| + | 5.80 real 11.19 user 0.00 sys | ||
| + | </pre> | ||
| + | |||
| + | This is the most accurate baseline comparison. With two threads, there are no slices. For absolute single-threaded execution, add -T to the flags. | ||
| + | |||
| + | To show the performance for additional threads, increase the number of jobs to the core count (4): | ||
| + | |||
| + | <pre> | ||
| + | $ time ./cmp_mp -l 12 -j 4 -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 8 | ||
| + | -l optarg = 12 | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 4, map_size = 14882 (0x3a22), slice_mod 0, against_sz 29764 | ||
| + | number of sequences: 223 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_4_12_0_100_nuc.csv | ||
| + | 2.91 real 11.21 user 0.00 sys | ||
| + | </pre> | ||
| + | |||
| + | A change to -j 16, the maximum thread-core count, yields | ||
| + | |||
| + | <pre> | ||
| + | $ time ./cmp_mp -l 12 -j 16 -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 8 | ||
| + | -l optarg = 12 | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 16, map_size = 3720 (0xe88), slice_mod 4, against_sz 29764 | ||
| + | number of sequences: 226 | ||
| + | longest sequence: 2845 ([19475, 19475), (22319, 22319)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_16_12_0_100_nuc.csv | ||
| + | 1.05 real 15.39 user 0.03 sys | ||
| + | </pre> | ||
| + | |||
| + | Notice the impact of slicing - there are more matching sequences, and the longest sequence is shorter. | ||
| + | |||
| + | There can be a minor performance benefit by double-loading the thread-core count: | ||
| + | |||
| + | <pre> | ||
| + | $ time ./cmp_mp -l 12 -j 32 -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 8 | ||
| + | -l optarg = 12 | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 32, map_size = 1860 (0x744), slice_mod 4, against_sz 29764 | ||
| + | thread[1] had no matches | ||
| + | number of sequences: 234 | ||
| + | longest sequence: 1860 ([13020, 13020), (14879, 14879)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_32_12_0_100_nuc.csv | ||
| + | 1.02 real 15.60 user 0.02 sys | ||
| + | </pre> | ||
| + | |||
| + | Post-processing output: | ||
| + | |||
| + | <center>[[file:Mp_CVD_OM003364.1_vs_CVD_OM002793.1_18_0_100_nuc.gif]]</center> | ||
| + | |||
| + | For completeness, the EBV against EBV comparison: | ||
| + | |||
| + | <center>[[file:Mp_EBV_KC207814.1_vs_EBV_NC_007605.1_12_0_100_nuc.gif]]</center> | ||
| + | |||
| + | It shows widely spread "repeat motifs" that are shared among different strains. The same (but different) sequences are found repeatedly within the whole organism's genomic sequence. The function of repeat motifs is not completely understood, but may provide a shape or sequence that helps evade host immune system responses. EBV is a member of the Herpes family, remains latent in B-cells, and can reactivate periodically, just like varicella zoster (Shingles/chickenpox) and oral and genital herpes simplex. | ||
| + | |||
| + | === pmcstat === | ||
| + | pmcstat is the standard for accessing Performance Monitor Counter (PMC) events with FreeBSD. Its syntax is relatively straightforward, with flags and the command with arguments, but there can be a bewhildering number of PMCs to measure. The man pages are helpful, and there is some built-in help. The [#Additional Resources] section has links to other discussions of how to use pmcstat. | ||
| + | |||
| + | <pre> | ||
| + | $ pmcstat -L | wc -l | ||
| + | 889 | ||
| + | </pre> | ||
| + | |||
| + | The -L flag lists all of the PMC events that can be monitored. Any entry does not guarantee operation. -u gives a short blurb for the entry: | ||
| + | |||
| + | <pre> | ||
| + | $ pmcstat -u pm_inst_cmpl | ||
| + | pm_inst_cmpl: Number of PowerPC Instructions that completed | ||
| + | </pre> | ||
| + | |||
| + | For many PMC events, pmcstat can be run without root privileges. | ||
| + | |||
| + | pmcstat can be set to monitor system-wide or application-specific monitoring. For this profiling application-specific will be used. While the application is executing, pmcstat samples the PMCs periodically. The default setting is -w 5 for five second output with per interval incrementing. Intervals can be fractions of a second. The flag -C enables cumulative counting. -v adds verbosity, but this does not always actually mean anything. For application sampling, -p is used to specify the PMC. It is used as often as needed. | ||
| + | |||
| + | A typical use to count the number of completed (not issued or canceled) instructions, but with sampling once per second: | ||
| + | |||
| + | <pre> | ||
| + | $ time pmcstat -w 1 -v -p pm_inst_cmpl ./cmp_mp -l 12 -j 2 -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 8 | ||
| + | -l optarg = 12 | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764 | ||
| + | # p/pm_inst_cmpl | ||
| + | 7521205 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 number of sequences: 222 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv | ||
| + | |||
| + | 973997 | ||
| + | 5.79 real 0.00 user 0.01 sys | ||
| + | </pre> | ||
| + | |||
| + | From the earlier discussion of the benchmark software, the output from the comparison code should be recognizable. Separating the pmcstat-specific output: | ||
| + | |||
| + | <pre> | ||
| + | # p/pm_inst_cmpl | ||
| + | 7521205 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | ... | ||
| + | 973997 | ||
| + | </code> | ||
| + | |||
| + | In the first second, 7,521,205 instructions completed. As can be seen, no additional instructions were completed, so the description is perhaps incomplete. This will be explored later. | ||
| + | |||
| + | A more extensive examination | ||
| + | |||
| + | <pre> | ||
| + | $ time pmcstat -w 1 -p pm_l1_icache_miss -p pm_ic_demand_cyc -p pm_ic_pref_req -p pm_inst_from_l1 ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | ... | ||
| + | # p/pm_l1_icache_miss p/pm_ic_demand_cyc p/pm_ic_pref_req p/pm_inst_from_l1 | ||
| + | 30081 589825 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | 1522 42563 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | 0 0 0 0 | ||
| + | ... | ||
| + | 2408 97726 0 0 | ||
| + | </pre> | ||
| + | |||
| + | This is looking closely at instruction cache misses, demands, prefetches and instructions that are already resident in the L1. | ||
| + | |||
| + | No load w/ smt=1 | ||
| + | |||
| + | <pre> | ||
| + | $ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 9 | ||
| + | -l optarg = 12 | ||
| + | multithreading disabled | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764 | ||
| + | # p/pm_l1_icache_miss | ||
| + | 30312 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 1459 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 number of sequences: 222 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv | ||
| + | |||
| + | 2399 | ||
| + | 11.18 real 0.01 user 0.00 sys | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | No load w/ smt=4 | ||
| + | |||
| + | <pre> | ||
| + | $ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 9 | ||
| + | -l optarg = 12 | ||
| + | multithreading disabled | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764 | ||
| + | # p/pm_l1_icache_miss | ||
| + | 47223 | ||
| + | 0 | ||
| + | 90369 | ||
| + | 0 | ||
| + | 90800 | ||
| + | 64188 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 number of sequences: 222 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv | ||
| + | |||
| + | 2411 | ||
| + | 11.18 real 0.01 user 0.00 sys | ||
| + | </pre> | ||
| + | |||
| + | Load w/ smt=1 | ||
| + | |||
| + | <pre> | ||
| + | $ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 9 | ||
| + | -l optarg = 12 | ||
| + | multithreading disabled | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764 | ||
| + | # p/pm_l1_icache_miss | ||
| + | 30449 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 1493 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 number of sequences: 222 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv | ||
| + | |||
| + | 2569 | ||
| + | 11.17 real 0.00 user 0.00 sys | ||
| + | </pre> | ||
| + | |||
| + | Load w/ smt=4 | ||
| + | |||
| + | <pre> | ||
| + | $ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1 | ||
| + | num args: 9 | ||
| + | -l optarg = 12 | ||
| + | multithreading disabled | ||
| + | input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta | ||
| + | o_st.st_size: 29766 , t_st.st_size: 29766 | ||
| + | min_pct_f = 1.00 | ||
| + | min_len = 12 | ||
| + | n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764 | ||
| + | # p/pm_l1_icache_miss | ||
| + | 61484 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 2348 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 | ||
| + | 0 number of sequences: 222 | ||
| + | longest sequence: 3270 ([19475, 19475), (22744, 22744)] | ||
| + | file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv | ||
| + | |||
| + | 7596 | ||
| + | 19.84 real 0.00 user 0.01 sys | ||
| + | </pre> | ||
| + | |||
| + | === perf === | ||
| + | |||
| + | === Additional Resources === | ||
| + | [[:File:POWER9 um OpenPOWER v21 10OCT2019 pub.pdf|POWER9 User Manual v21]] | ||
| + | |||
| + | [[:File:POWER9_PMU_UG_v12_28NOV2018_pub.pdf|POWER9 Performance Monitoring Unit User Guide v12]] | ||
| + | |||
| + | [https://www.ibm.com/support/pages/power-cpu-memory-affinity-3-scheduling-processes-smt-and-virtual-processors POWER CPU Memory Affinity 3 - Scheduling processes to SMT and Virtual Processors] | ||
| + | |||
| + | https://www.ibm.com/docs/en/linux-on-systems?topic=linuxonibm/performance/tuneforsybase/smtsettings.htm | ||
| + | |||
| + | [https://freebsdfoundation.org/wp-content/uploads/2014/03/Understanding-Application-and-System-Performance-with-HWPMC4.pdf George Neville-Neil's brief tutorial on pmcstat] | ||
Latest revision as of 13:08, 16 July 2025
This article discusses profiling symmetric multithreading (SMT) on the POWER9 architecture. It uses both Big-endian FreeBSD with pmcstat and Debian with perf.
The knowledge presented here was derived from a variety of sources which can be found in the #Additional Resources section.
Contents
Symmetric multithreading (SMT)
SMT principles
SMT is "multi-thread" not "multi-core"
This is an important distinction. SMT is a technology that increases throughput of instructions through parallelization where there are under-used CPU components. While SMT4 can support four threads per core and SMT8 can support eight threads per core, this is not an additional three and seven cores, respectively. There are trade-offs and benefits. Per-thread performance declines with increasing utilization of SMT levels, but overall performance and power consumption efficiency increase. Note that IBM did not market SMT as "multi-core," while several media sites conflated SMT with increased core count.
Comparison to RISC-V HARTs
Operating Environment
Hardware
The bare metal hardware is an eight core SMT4-capable system from Raptor.
# lscpu Architecture: ppc64le Byte Order: Little Endian CPU(s): 32 On-line CPU(s) list: 0-31 Model name: POWER9 (raw), altivec supported Model: 2.3 (pvr 004e 1203) Thread(s) per core: 4 Core(s) per socket: 8 Socket(s): 1 Frequency boost: enabled CPU(s) scaling MHz: 67% CPU max MHz: 3800.0000 CPU min MHz: 2166.0000 Caches (sum of all): L1d: 256 KiB (8 instances) L1i: 256 KiB (8 instances) NUMA: NUMA node(s): 1 NUMA node0 CPU(s): 0-31 Vulnerabilities: Gather data sampling: Not affected Indirect target selection: Not affected Itlb multihit: Not affected L1tf: Mitigation; RFI Flush, L1D private per thread Mds: Not affected Meltdown: Mitigation; RFI Flush, L1D private per thread Mmio stale data: Not affected Reg file data sampling: Not affected Retbleed: Not affected Spec rstack overflow: Not affected Spec store bypass: Mitigation; Kernel entry/exit barrier (eieio) Spectre v1: Mitigation; __user pointer sanitization, ori31 speculation barrier enabled Spectre v2: Mitigation; Software count cache flush (hardware accelerated), Software link stack flush Srbds: Not affected Tsx async abort: Not affected # lsmem RANGE SIZE STATE REMOVABLE BLOCK 0x0000000000000000-0x00000007ffffffff 32G online yes 0-31 Memory block size: 1G Total online memory: 32G Total offline memory: 0B # facter | grep virtual is_virtual => false virtual => physical
Software
Three environments will be used for the operating environment. The bare metal runs Debian:
# uname -a Linux 93-224-155-23 6.1.0-37-powerpc64le #1 SMP Debian 6.1.140-1 (2025-05-22) ppc64le GNU/Linux
Two QEMU VMs provide little-endian Debian and big-endian FreeBSD environments:
# lscpu Architecture: ppc64le Byte Order: Little Endian CPU(s): 16 On-line CPU(s) list: 0,4,8,12 Off-line CPU(s) list: 1-3,5-7,9-11,13-15 Model name: POWER9 (architected), altivec supported Model: 2.3 (pvr 004e 1203) Thread(s) per core: 1 Core(s) per socket: 4 Socket(s): 1 Virtualization features: Hypervisor vendor: KVM Virtualization type: para Caches (sum of all): L1d: 128 KiB (4 instances) L1i: 128 KiB (4 instances) NUMA: NUMA node(s): 1 NUMA node0 CPU(s): 0,4,8,12 Vulnerabilities: Gather data sampling: Not affected Indirect target selection: Not affected Itlb multihit: Not affected L1tf: Mitigation; RFI Flush, L1D private per thread Mds: Not affected Meltdown: Mitigation; RFI Flush, L1D private per thread Mmio stale data: Not affected Reg file data sampling: Not affected Retbleed: Not affected Spec rstack overflow: Not affected Spec store bypass: Mitigation; Kernel entry/exit barrier (eieio) Spectre v1: Mitigation; __user pointer sanitization, ori31 speculation barrier enabled Spectre v2: Mitigation; Software count cache flush (hardware accelerated), Software link stack flush Srbds: Not affected Tsx async abort: Not affected # lsmem RANGE SIZE STATE REMOVABLE BLOCK 0x0000000000000000-0x00000001ffffffff 8G online yes 0-511 Memory block size: 16M Total online memory: 8G Total offline memory: 0B
Benchmark code
Description of comparison code (cmp_mp)
The code being profiled and used for benchmarking is the genomic comparison code that the M. P. Janson Institute for Analytical Medicine uses to look for molecular mimicry between pathogens and human tissue and hormones. This code uses a variety of techniques to compare nucleotide and amino acid sequences. There are both byte-by-byte and vector versions. The raw source code can be found at TBD
Fundamentally the algorithm just compares characters in two sequences for matching. The goal is to match consecutive characters between two sequences with constantly changing characters, and record the string and location of the matches. Unlike strstr or similar functions, the string to be matched is not known in advance. The first character in one sequence is compared against the first character in the other sequence. If it matches, the second characters are compared. If they match, the third characters are compared, and so on, until the characters do not match. If the length of the matched string is below the minimum desired length, the match is discarded and the comparisons start over at the next character. This can "slide" the comparisons relative to each other, so if the first four characters match but not the fifth, the comparison starts over at the first character of one sequence and the fifth character of the other. If there is a match of sufficient length, that two-dimensional region is marked as an "exclusion region" and not searched again.
The code can run N number of user-specified pthreads in parallel (-j N), by slicing one of the two sequences into smaller pieces and stitching the results together. N must be an even number (one forward, one reverse). Some matches are lost at the boundaries between slices when one or both parts are less than the minimum length. The execution time is extremely sensitive to comparisons made; adding checks to determine if a match falls on a slice boundary adds significant overhead. For large sequences, the odds of a matching string falling on a boundary are very low; for small sequences, running fewer threads is effective without costing significant time delays. For absolute accuracy, run only two threads (one forward and one in reverse).
Description of data
Nucleotides are denoted as A, T, C, and G. There are three nucleotides in an amino acid, and 23 amino acids in humans, which leads to "isocodon" amino acids. These have the same function but different nucleotide combinations. There are amino acids that denote the initiation and termination of a gene. The amino acid is determined by its offset from the initiation amino acid (ATG, methionine), which starts an "Open Reading Frame (ORF)." ORFs can overlap. DNA has a forward ("sense") and backward ("anti-sense") direction, so there are three possible amino acids reading forward, and three when reading backwards. Nucleotide sequences are usually published in the forward direction, and the other strand's nucleotide matching the forward direction is inferred (A matches T, and C matches G, and vice versa). Genome sequences can be obtained from the NIH National Library of Medicine gene database, as well as in other places. The particular sequence is identified by its "accession number," and sequencing of the same organism by different labs does lead to different accession numbers. Some sequences are "canonical," meaning the generally accepted sequence that is often an assembly of accessions, and others are "contributions," meaning independently obtains sequences submitted for recording.
A typical "FASTA" file contains data as
TATATTAGAGTAGGAGCTAGAAAATCAGCACCTTTAATTGAATTGTGCGTGGATGAGGCTGGTTCTAAATCACCCATTCAGTACATCGATATCGGTAATTATACAGTT...
The most common comparison method is BLAST, which uses a database of pre-identified nucleotide sequences. This is useful for rapidly finding large matches, but does not offer granularity for exploring other aspects of the genome. The above method allows matching of nucleotide and amino acid sequences on both position one and position two nucleotide. This often reveals "repeat motifs" where a sequence of nucleotides is repeated multiple times within a pathogen. The current thinking is that this is an "immuno-evasive strategy," allowing the pathogen to escape detection by the host's immune system. Additionally, by recording the location of the matches, the distance between matches can be calculated. This is important in deciding if a host antibody would identify healthy tissue and hormones as pathogenic, by having similar shapes. The above algorithm also allows for detecting "SIDs," which are "Substitutions, Insertions, and Deletions," which occur regularly in rapidly evolving pathogens such as viruses.
For example, relative to the example above
TTATATTAGAGTAGGAGCTAGAAAATCAGCACCTTTAATTGAATTGTGCGTGGATGAGGCTGGTTCTAAATCACCCATTCAGTACATCGATATCGGTAATTATACAGTT...
is identical except for the inserted nucleotide T at the very first character. If the ORF is prior to the sequence above, this changes the amino acid structure by altering the anchor point for examining three nucleotides at a time (a "codon"). A straight matching of amino acids would miss this. The comparison code (cmp_mp) has the ability to shift each nucleotide sequence by none, one and two positions to catch insertions that alter the identifiable amino acids, and can match against the first character ("T" in "TAA") or the second nucleotide ("A" in "TAA"), which is more highly conserved across evolutionary pathways.
Sample output
The output is a csv file:
CVD_OM003364.1,CVD_OM002793.1,225,2848,29762,29762,1,1, 1,1,163,163,164, ...
The first line indicates the sequences compared, the number of matches, the longest match, the length of each sequence, and the presentation form, in this case a 1x1 image. The second line indicates a match was found from (1, 163) in one sequence and (1, 163) in the other, for a length of 164 characters, inclusive.
The csv file is run through some custom image generation code using libjpeg. An example of the processed data is below
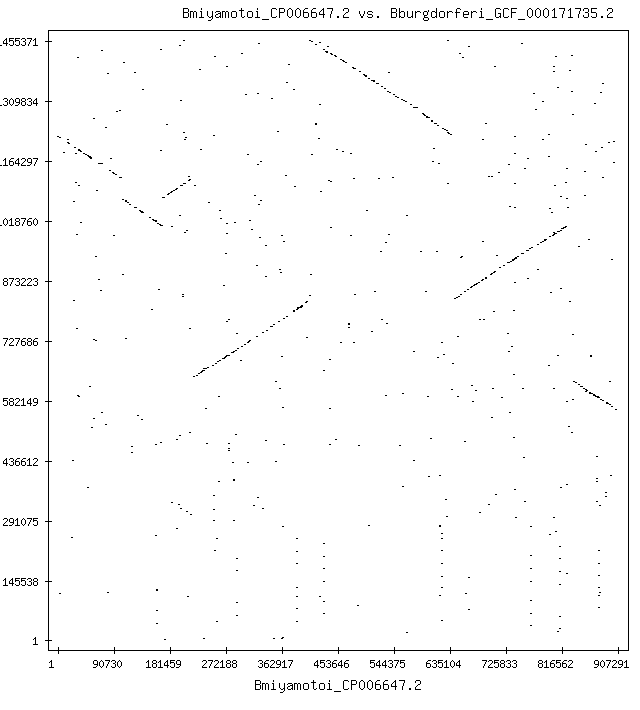
The image shows matching sequences between two bacteria, B. burgdorferi, which causes Lyme disease, and B. miyamotoi, a bacteria in the same Borrelia family. Matches are occurring in both the forward and reverse directions. In this particular case, the B. miyamotoi sequence was assembled from fragments in both the "sense" and "antisense" direction. This is an artifact of how PCR is done, but has to be considered when doing analysis. Running only forward matching can lead to missed matches (a "false negative"). This comparison is particularly important because some people will exhibit symptoms of Lyme disease but only test weakly positive or not at all for Lyme disease. This is known in science but not practiced in medicine, leading to erroneous conclusions about the cause of the patient's symptoms (see Borrelia miyamotoi infection leads to cross-reactive antibodies to the C6 peptide in mice and men).
The B. burgdefori sequence is 1,455,375 nucleotides long, and the B. miyamotoi sequence is 907,293 nucleotides long. Each nucleotide is compared at least once, and when there are partial matches too short to place the nucleotide into an exclusion region, it may be compared more than once. In this case, more than 2.6 trillion comparisons are made (one comparison is forward against forward, and one is forward against reverse; reverse against reverse is the same as forward against forward because of the inverse nucleotide arrangement of DNA). When searching human genes, the comparisons can be on the order of quadrillions (10^15) and take hours even on fast CPUs even with multi-core threading. Optimizing the comparison code saves time and power consumption, and is the focus of the profiling done below.
Baseline comparison
For these benchmarks, two different sequences of Epstein-Barr (EBV) and SARS-CoV-2 (Covid-19) virii will be used. EBV is the virus that causes mononucleosis, but is also closely associated with multiple sclerosis and has identifiable molecular mimicry sequences. SARS-CoV-2, the virus that causes Covid-19, evolved from a coronavirus, as can be shown by a comparison of amino acids (nucleotide shifts of none for CRNA and one for Covid-19, anchor on second position, match initiation, termination and arginine amino acids):
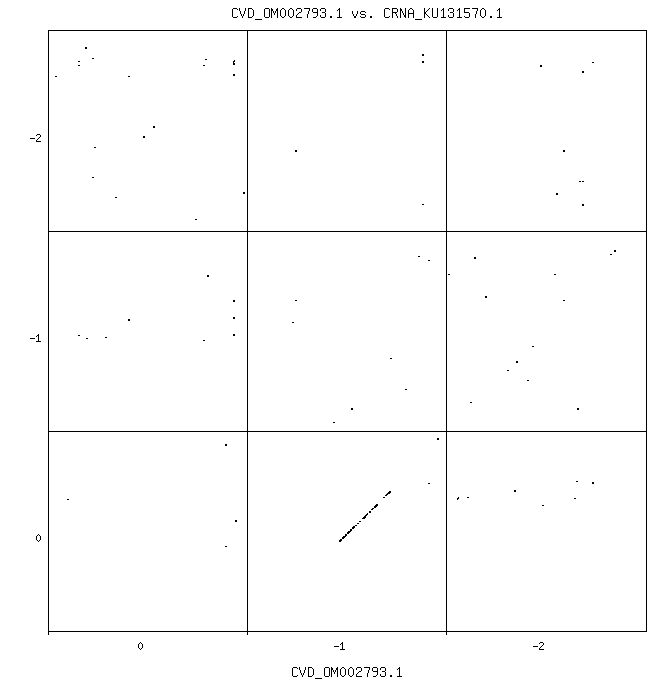
The COVID-19 sequences, OM002793.1 and OM003364.1 are suitable for short duration benchmarks. The EBV sequences, KC207814.1 and NC_007605.1 (canonical), are for longer benchmarks in which slight performance gains are measured. Each pair member in the two pairs of sequences is nearly identical to the other, so when analysis of branch prediction for misses is needed, one from each pair can be compared against the other.
A typical comparison command looks like
./cmp_mp -l 12 -j 8 -s CVD_OM002793.1 CVD_OM003364.1
It is specifying a minimum length of 12 consecutive nucleotide matches and four forward slices and four reverse slices, with the two Covid-19 sequences. For benchmark clarity, the -s (silence) flag is set. The output from BE FreeBSD for only two threads:
$ time ./cmp_mp -l 12 -j 1 -s CVD_OM002793.1 CVD_OM003364.1
num args: 8
-l optarg = 12
Number of jobs must be even; adding +1 to n_jobs
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764
number of sequences: 222
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv
5.80 real 11.19 user 0.00 sys
This is the most accurate baseline comparison. With two threads, there are no slices. For absolute single-threaded execution, add -T to the flags.
To show the performance for additional threads, increase the number of jobs to the core count (4):
$ time ./cmp_mp -l 12 -j 4 -s CVD_OM002793.1 CVD_OM003364.1
num args: 8
-l optarg = 12
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 4, map_size = 14882 (0x3a22), slice_mod 0, against_sz 29764
number of sequences: 223
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_4_12_0_100_nuc.csv
2.91 real 11.21 user 0.00 sys
A change to -j 16, the maximum thread-core count, yields
$ time ./cmp_mp -l 12 -j 16 -s CVD_OM002793.1 CVD_OM003364.1
num args: 8
-l optarg = 12
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 16, map_size = 3720 (0xe88), slice_mod 4, against_sz 29764
number of sequences: 226
longest sequence: 2845 ([19475, 19475), (22319, 22319)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_16_12_0_100_nuc.csv
1.05 real 15.39 user 0.03 sys
Notice the impact of slicing - there are more matching sequences, and the longest sequence is shorter.
There can be a minor performance benefit by double-loading the thread-core count:
$ time ./cmp_mp -l 12 -j 32 -s CVD_OM002793.1 CVD_OM003364.1
num args: 8
-l optarg = 12
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 32, map_size = 1860 (0x744), slice_mod 4, against_sz 29764
thread[1] had no matches
number of sequences: 234
longest sequence: 1860 ([13020, 13020), (14879, 14879)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_32_12_0_100_nuc.csv
1.02 real 15.60 user 0.02 sys
Post-processing output:
For completeness, the EBV against EBV comparison:
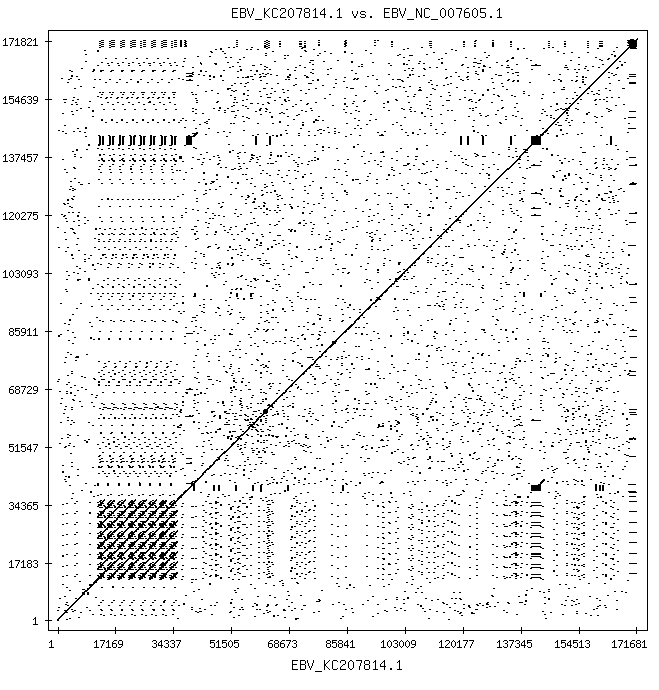
It shows widely spread "repeat motifs" that are shared among different strains. The same (but different) sequences are found repeatedly within the whole organism's genomic sequence. The function of repeat motifs is not completely understood, but may provide a shape or sequence that helps evade host immune system responses. EBV is a member of the Herpes family, remains latent in B-cells, and can reactivate periodically, just like varicella zoster (Shingles/chickenpox) and oral and genital herpes simplex.
pmcstat
pmcstat is the standard for accessing Performance Monitor Counter (PMC) events with FreeBSD. Its syntax is relatively straightforward, with flags and the command with arguments, but there can be a bewhildering number of PMCs to measure. The man pages are helpful, and there is some built-in help. The [#Additional Resources] section has links to other discussions of how to use pmcstat.
$ pmcstat -L | wc -l
889
The -L flag lists all of the PMC events that can be monitored. Any entry does not guarantee operation. -u gives a short blurb for the entry:
$ pmcstat -u pm_inst_cmpl pm_inst_cmpl: Number of PowerPC Instructions that completed
For many PMC events, pmcstat can be run without root privileges.
pmcstat can be set to monitor system-wide or application-specific monitoring. For this profiling application-specific will be used. While the application is executing, pmcstat samples the PMCs periodically. The default setting is -w 5 for five second output with per interval incrementing. Intervals can be fractions of a second. The flag -C enables cumulative counting. -v adds verbosity, but this does not always actually mean anything. For application sampling, -p is used to specify the PMC. It is used as often as needed.
A typical use to count the number of completed (not issued or canceled) instructions, but with sampling once per second:
$ time pmcstat -w 1 -v -p pm_inst_cmpl ./cmp_mp -l 12 -j 2 -s CVD_OM002793.1 CVD_OM003364.1
num args: 8
-l optarg = 12
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764
# p/pm_inst_cmpl
7521205
0
0
0
0 number of sequences: 222
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv
973997
5.79 real 0.00 user 0.01 sys
From the earlier discussion of the benchmark software, the output from the comparison code should be recognizable. Separating the pmcstat-specific output:
# p/pm_inst_cmpl
7521205
0
0
0
0
...
973997
</code>
In the first second, 7,521,205 instructions completed. As can be seen, no additional instructions were completed, so the description is perhaps incomplete. This will be explored later.
A more extensive examination
<pre>
$ time pmcstat -w 1 -p pm_l1_icache_miss -p pm_ic_demand_cyc -p pm_ic_pref_req -p pm_inst_from_l1 ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1
...
# p/pm_l1_icache_miss p/pm_ic_demand_cyc p/pm_ic_pref_req p/pm_inst_from_l1
30081 589825 0 0
0 0 0 0
0 0 0 0
0 0 0 0
0 0 0 0
1522 42563 0 0
0 0 0 0
0 0 0 0
0 0 0 0
0 0 0 0
0 0 0 0
...
2408 97726 0 0
This is looking closely at instruction cache misses, demands, prefetches and instructions that are already resident in the L1.
No load w/ smt=1
$ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1
num args: 9
-l optarg = 12
multithreading disabled
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764
# p/pm_l1_icache_miss
30312
0
0
0
0
1459
0
0
0
0
0 number of sequences: 222
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv
2399
11.18 real 0.01 user 0.00 sys
No load w/ smt=4
$ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1
num args: 9
-l optarg = 12
multithreading disabled
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764
# p/pm_l1_icache_miss
47223
0
90369
0
90800
64188
0
0
0
0
0 number of sequences: 222
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv
2411
11.18 real 0.01 user 0.00 sys
Load w/ smt=1
$ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1
num args: 9
-l optarg = 12
multithreading disabled
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764
# p/pm_l1_icache_miss
30449
0
0
0
0
1493
0
0
0
0
0 number of sequences: 222
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv
2569
11.17 real 0.00 user 0.00 sys
Load w/ smt=4
$ time pmcstat -w 1 -p pm_l1_icache_miss ./cmp_mp -l 12 -j 2 -T -s CVD_OM002793.1 CVD_OM003364.1
num args: 9
-l optarg = 12
multithreading disabled
input/CVD_OM002793.1.fasta, input/CVD_OM003364.1.fasta
o_st.st_size: 29766 , t_st.st_size: 29766
min_pct_f = 1.00
min_len = 12
n_jobs = 2, map_size = 29764 (0x7444), slice_mod 0, against_sz 29764
# p/pm_l1_icache_miss
61484
0
0
0
0
0
0
0
0
2348
0
0
0
0
0
0
0
0
0 number of sequences: 222
longest sequence: 3270 ([19475, 19475), (22744, 22744)]
file name: mp_CVD_OM003364.1_vs_CVD_OM002793.1_2_12_0_100_nuc.csv
7596
19.84 real 0.00 user 0.01 sys
perf
Additional Resources
POWER9 Performance Monitoring Unit User Guide v12
POWER CPU Memory Affinity 3 - Scheduling processes to SMT and Virtual Processors